ABSTRACT
In silico prediction of antileishmanial activity using quantitative structure–activity relationship (QSAR) models has been developed on limited and small datasets. Nowadays, the availability of large and diverse high-throughput screening data provides an opportunity to the scientific community to model this activity from the chemical structure. In this study, we present the first KNIME automated workflow to modeling a large, diverse, and highly imbalanced dataset of compounds with antileishmanial activity. Because the data is strongly biased toward inactive compounds, a novel strategy was implemented based on the selection of different balanced training sets and a further consensus model using single decision trees as the base model and three criteria for output combinations. The decision tree consensus was adopted after comparing its classification performance to consensuses built upon Gaussian-Naı̈ve-Bayes, Support-Vector-Machine, Random-Forest, Gradient-Boost, and Multi-Layer-Perceptron base models. All these consensuses were rigorously validated using internal and external test validation sets and were compared against each other using Friedman and Bonferroni–Dunn statistics. For the retained decision tree-based consensus model, which covers 100% of the chemical space of the dataset and with the lowest consensus level, the overall accuracy statistics for test and external sets were between 71 and 74% and 71 and 76%, respectively, while for a reduced chemical space (21%) and with an incremental consensus level, the accuracy statistics were substantially improved with values for the test and external sets between 86 and 92% and 88 and 92%, respectively. These results highlight the relevance of the consensus model to prioritize a relatively small set of active compounds with high prediction sensitivity using the Incremental Consensus at high level values or to predict as many compounds as possible, lowering the level of Incremental Consensus. Finally, the workflow developed eliminates human bias, improves the procedure reproducibility, and allows other researchers to reproduce our design and use it in their own QSAR problems.
AUTHORS
Omar Casanova-Alvarez∥, Aliuska Morales-Helguera†, Miguel Ángel Cabrera-Pérez†, Reinaldo Molina-Ruiz†, and Christophe Molina*
∥Departamento de Química, Facultad de Química-Farmacia, Universidad Central “Marta Abreu” de Las Villas, Santa Clara, Villa Clara 54830, Cuba
†Centro de Bioactivos Químicos. Universidad Central “Marta Abreu” de las Villas. Santa Clara 54830, Villa Clara, Cuba
*PIKAÏROS S.A, 31650 Saint Orens de Gameville, France
JOURNAL
Journal of Chemical Information and Modeling ( https://doi.org/10.1021/acs.jcim.0c01439)
SUPPORTING INFORMATION
How to install and start with KNIME : https://www.knime.com/knime
KNIME WORKFLOW : Available at https://pikairos.eu/download/leishmania_classification (3.03 GBytes)
Please get in touch with us through our Contact Page if you have any questions concerning the journal article or the KNIME Workflow.
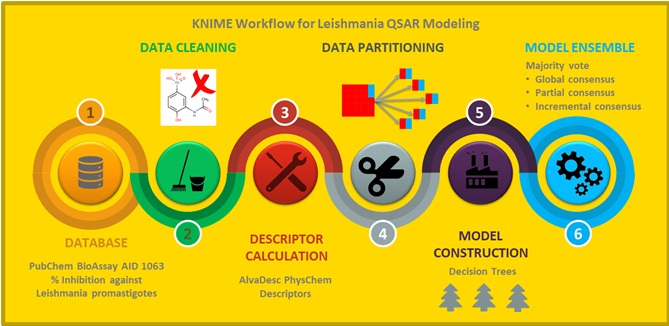
Workflow Available under the Revised BSD License
Copyright (c) 2020-2025, Omar Casanova-Alvarez, Aliuska Morales-Helguera, Miguel Ángel Cabrera-Pérez, Reinaldo Molina-Ruiz, Christophe Molina & Pikaïros.
All rights reserved. Redistribution and use in source and binary forms of this workflow, with or without modification, are permitted provided that the following conditions are met:
- Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
- Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
- Neither the name of Pikaïros nor the names of its contributors may be used to endorse or promote products derived from this software without specific prior written permission.
THIS WORKFLOW IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS “AS IS” AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE AUTHORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
